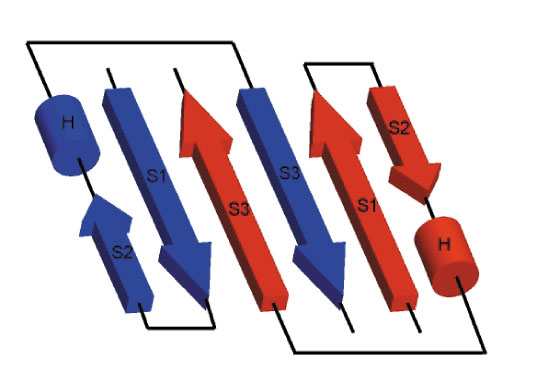
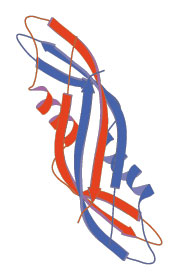
........10........20........30........40........50........60 MIMTSMTLDLPRRFPWPTLLSVCIHGAVVAGLLYTSVHQVIELPAPAQPISVTMVTPADL ---EEEE-----------EEEEEHHHHHHHHHEEEEEEEEE--------EEEEEE----- PHD 964234167788687883378722345541112222221322799999679998427899 CCEEEEEECCCCCCCCCEEEEECHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCCCCCCC PSIpred 920122210235557423442200278888899871363104678854687122687656 -----------------EEEEEE--HHHHHHHHHH----EE--------EEEEEE----- Prof 976544445677788864687644567888877754444446788998758998657766 ---------------HHHHHHHHHHHHHHHHHHHH-HHHHH--------EEEEEE----- SSpro ------------------------------------------------------------ JPred NNNNN??????????????????????????????????????????????????????? 3D ........70........80........90.......100.......110.......120 EPPQAVQPPPEPVVEPEPEPEPIPEPPKEAPVVIEKPKPKPKPKPKPVKKVQEQPKRDVK ------------------------------------------------------------ PHD 999999999999999999998999999999732368999999999989886768898989 ------------------------------------------------HHHH-------- PSIpred 787788888688708645556778787878866788456778887010221001224432 -------------------------------EEEE------------------------- Prof 656565555554445555555555555544445444445555556555554456778889 --------------------------------EE------------------------E- SSpro ------------------------------------------------------------ JPred ???????????????????????????????????????????????????????????? 3D .......130.......140.......150.......160.......170.......180 PVESRPASPFENTAPARLTSSTATAATSKPVTSVASGPRALSRNQPQYPARAQALRIEGQ -------------------------------------------------HHHHHHH---- PHD 999999999899998967898898899999988779986535899998168886523465 --------------------------------HHHH---HHHH---HHHHHHHH----EE PSIpred 013578878888774555777343888785661001143144578611378873575228 ------------------------------------------------HHHHHHH----E Prof 899999999998887655554433333445444678886557898888578998747867 ------------------------------------------------HHHHHH----EE SSpro ------------------------------------------------------------ JPred ????????????????????????????????????????????????-----EEEEEEE 3D .......190.......200.......210.......220.......230.......240 VKVKFDVTPDGRVDNVQILSAKPANMFEREVKNAMRRWRYEPGKPGSGIVVNILFKINGT EEEEEEE------EEEEEEE---HHHHHHHHHHHHHHHHH--------EEEEEEEEE--- PHD 599998389995323697504981489999999999975618989981579999983694 EEEEEEE----EEEEEEEE------HHHHHHHHHHHH-----------EEEEEEEEE--- PSIpred 899999899971889998526897067699999997064868888787269999997771 EEEEEEEE------EEEEEE----HHHHHHHHHHHHHHHH--------EEEEEEEEE--- Prof 999999857888657899856885578999999999987578988887689999985677 EEEEEEE--------EEEEE---HHHHHHHHHHHHHH------------EEEEEEEE--- SSpro ----------------EE-----------HHHHH-------------------EEE---- JPred EEEEEEE-----EEEEEEE-----HHHHHHHHHHHh---------EEEEEEEEEEEEEEE 3D TEIQ -E-- PHD 2149 ---- PSIpred 0279 ---- Prof 6679 EE-- SSpro ---- JPred E--- 3D
Latest update of content: June 19, 2003 Ralf Koebnik
| ||||||||||||||||||||||||