|
Prediction of coiled-coil regions in proteins:
Having solved many 3D structures at atomic resolution one realized that successive secondary structural elements (alpha-helices and beta-strands) assemble into typical structural motifs, called super-secondary structure (i.e. a form of structural organization between secondary and der tertiary structure).
Examples of such motifs are beta-hairpins and helix-turn-helix motifs.
Even more complex associations of secondary structural elements are called (domain) folds, e.g. the Rossmann fold, Greek key and four-helix bundle.

Coiled coils are a special form of associated secondary structural elements.
In this case, two, three, four or five alpha-helices associate with each other in a parallel or anti-parallel manner.
Thereby the alpha-helices can belong to only one or also several (identical or different) proteins.
Thus, coiled-coils can promote the formation of homo- or heterooligomeric protein complexes.
Due to the fact that ca. 3 % of all proteins contain coiled-coil domains, this form of protein-protein interaction is probably important for many cellular processes.
The lab of Peter S. Kim and Bonnie Berger at the MIT in Boston used structural and computational biology, biochemistry as well as genetics to decipher the "code" that determines the specificity and stability of coiled-coil interactions.
This knowledge could be used to predict protein-protein interactions at the proteome scale.
Coiled coils are characterized by a repetition of a short sequence motif, the so-called heptad repeat, which usually contains hydrophobic amino acids at the first and fourth position (a und d) of the repeat.
The following papers provide a good overview:
Andrei Lupas (1996)
Coiled-coils: New structures and new functions
Trends Biochem. Sci. 21: 375-382
Andrei Lupas (1997)
Predicting coiled-coil regions in proteins
Curr. Opin. Struct. Biol. 7: 388-393
Peter Burkhard, Jörg Stetefeld, and Sergei V. Strelkov (2001)
Coiled coils: a highly versatile protein folding motif
Trends Cell Biol. 11: 82-88
In the course we will get to know two algorithms which are based on statistical analyses:
- The program COILS
 was developed to predict left-handed coiled coils.
The corresponding documentation provides details for the choice of parameters,
such as two different matrices which can be used in a weighted (stronger weight of the a and d position within the heptad repeat) or unweighted manner. was developed to predict left-handed coiled coils.
The corresponding documentation provides details for the choice of parameters,
such as two different matrices which can be used in a weighted (stronger weight of the a and d position within the heptad repeat) or unweighted manner.
A. Lupas, M. Van Dyke, and J. Stock (1991)
Predicting coiled coils from protein sequences
Science 252: 1162-1164
- The program PairCoil
 was developed to predict two-stranded parallel coiled coils. was developed to predict two-stranded parallel coiled coils.
Bonnie Berger, David B. Wilson, Ethan Wolf, Theodore Tonchev, Maria Milla, and Peter S. Kim (1995)
Predicting coiled coils by use of pairwise residue correlations
Proc. Natl. Acad. Sci. USA 92: 8259-8263
- The subsequent program MultiCoil
 allows to predict and distinguish two-stranded and three-stranded coiled coils. allows to predict and distinguish two-stranded and three-stranded coiled coils.
Ethan Wolf, Peter S. Kim, and Bonnie Berger (1997)
MultiCoil: A program for predicting two- and three-stranded coiled coils
Protein Science 6: 1179-1189
- Later, specific programs were developed to analyze candidate coiled-coil structures with certain protein families.
The program LearnCoil-HK can be used to analyze histidine kinase receptors.
Mona Singh, Bonnie Berger, Peter S. Kim, James M. Berger, and Andrea G. Cochran (1998)
Computational learning reveals coiled coil-like motifs in histidine kinase linker domains
Proc. Natl. Acad. Sci. USA 95: 2738-2743
- LearnCoil-VMF can be used to identify coiled-coil structures in viral membrane fusion proteins, including those of retroviruses, paramyxoviruses and filoviruses.
Mona Singh, Bonnie Berger, and Peter S. Kim (1999)
LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins
J. Mol. Biol. 290: 1031-1041
- Alternatively, a hidden Markov model, Marcoil, was generated to predict coiled coils.

Mauro Delorenzi and Terry Speed (2002)
An HMM model for coiled-coil domains and a comparison with PSSM-based predictions
Bioinformatics 18: 617-625
- Another webserver using a hidden Markov model, CCHMM, can be used at the University of Bologna.

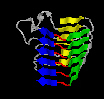
Another specific form of association of secondary structural elements is the parallel beta-helix.
Here, three successive beta-strands form kind of a triangle, several of which stack on each other and thus form three long beta-sheets.
These sequence modules contain typical amino acid sequences which allow to predict them.
By way of comparison to the coiled coils, this structural motif is rare and mainly restricted to secreted proteins.
- The program BETAWRAP allows to predict beta-helices.
Phil Bradley, Lenore Cowen, Matthew Menke, Jonathan King, and Bonnie Berger (2001)
BETAWRAP: successful prediction of parallel beta -helices from primary sequence reveals an association with many microbial pathogens
Proc. Natl. Acad. Sci. USA 98: 14819-14824
Sequence examples:
- Sucrose-Porin ScrY from Salmonella typhimurium
- Maltoporin LamB from Salmonella typhimurium
- Braun'sches Murein-Lipoprotein Lpp from Escherichia coli
- Phage shock protein PspA from Escherichia coli
- Pectate Lyase C PelC from Erwinia chrysanthemi
- Pertactin P.69 from Bordetella pertussis
- Tailspike protein gp9 from the Salmonella-specific bacteriophage P22
Latest update of content: December 19, 2005
Ralf Koebnik
Institut de recherche pour le dèveloppement
UMR 5096, CNRS-UP-IRD
911, Avenue Agropolis, BP 64501
34394 Montpellier, Cedex 5
FRANCE
Phone: +33 (0)4 67 41 62 28
Fax: +33 (0)4 67 41 61 81
Email: koebnik(at)gmx.de
Please replace (at) by @.
 Back to main page
Back to main page
|



 was developed to predict left-handed coiled coils.
The corresponding
was developed to predict left-handed coiled coils.
The corresponding  was developed to predict two-stranded parallel coiled coils.
was developed to predict two-stranded parallel coiled coils.
 allows to predict and distinguish two-stranded and three-stranded coiled coils.
allows to predict and distinguish two-stranded and three-stranded coiled coils.
